GATE COURSE MODULE 11
GATE & PYTHON
GATE & PYTHON
- Online slides: https://gatenlp.github.io/python-gatenlp/training/module11-python.slides.html
- Slides License: CC BY-NC-SA 3.0
GATE & PYTHON¶
This tutorial covers Python tools related to GATE:
- Python GateNLP: Python package for NLP similar to Java GATE
- Python GateNLP GateWorker: run Java/GATE from Python
- GATE Python Plugin: Java GATE plugin to process GATE documents with Python and Python GateNLP
- Format BDOC Plugin: Java GATE plugin for support of loading/saving documents in JSON/YAML/MsgPack format
Python GateNLP¶
Aims:
- NLP framework written in pure Python.
- Similar concepts as Java GATE: documents, document features, annotation sets, annotations, ...
- But "pythonic" API, try to make basic things very simple (e.g. loading/saving of documents)
- Does NOT try to be a full multilingual NLP processing package, rather COMBINE:
- Use existing tools and solutions: Spacy, Stanford Stanza
- Add own tools and improvements where needed
Python GateNLP: status¶
- Current release: 1.0.x
- All 1.0.x: get community feedback:
- how to improve API, abstractions, conventions, find bugs
- what is most important to still get added?
- API may slightly change, parameter names may get consolidated
- Planned 1.1.x releases and onwards: stable API
Python GateNLP: Info and Feedback¶
- Documentation: https://gatenlp.github.io/python-gatenlp/
- Sources: https://github.com/GateNLP/python-gatenlp
- Report a bug, request a feature with issue tracker: https://github.com/GateNLP/python-gatenlp/issues
- Discuss, ask:
- discussions forum at https://github.com/GateNLP/python-gatenlp/discussions
- GATE mailing list https://groups.io/g/gate-users
- Developers Chat: https://gitter.im/GateNLP/python-gatenlp
Preparation: Install Python¶
- see also https://gatenlp.github.io/python-gatenlp/installation.html
- Recommended:
Preparation: install Miniconda (Windows)¶
- Download the Python 3.8 (or later) installer (64-bit) for your OS
- Run the installer, install for "just me", register as default Python,
- start the "Anaconda Prompt" or "Anaconda Powershell Prompt"
- Create environment:
conda create -y -n gatenlp python=3.9 - activate environment:
conda activate gatenlp
Install gatenlp¶
To install most recent release and install all dependencies
(without [all] only minimum dependencies are installed!):
pip install -U gatenlp[all]
Also install support for jupyter notebookd and for showing the slides:
pip install jupyter notebook ipython ipykernel RISE
Create kernel for the conda environment:
python -m ipykernel install --user --name gatenlp --display-name "Python-gatenlp"
See also GateNLP documentation: installation
Follow along¶
- Online slides
- Download the handouts zip file and extract the directory
- Within the directory you can either:
- to follow in the original notebook: run
jupyter notebook module11-python.ipynb - to explore in a new Notebook: run
jupyter notebook, choose New -> Python-gatenlp - to explore interactively: run
ipythonand enter python code
- to follow in the original notebook: run
If kernel error in Jupyter, try something like (Anaconda bug, apparently):
python C:\Users\USERNAME\miniconda3\envs\gatenlp\Scripts\pywin32_postinstall.py -install
Python GateNLP: Main Concepts¶
- A document represents some text and
- any number of named annotation sets
- any number of features
- An annotation set can have
- any number of annotations
- Annotations describe a span of a document and have
- any number of features
- an annotation type
- from and to offsets that describe the span of the annotation
- An Annotator is something that processes a document (and usually adds or changes annotations)
Documents¶
# Import gatenlp to check gatenlp version:
import gatenlp
print("GateNLP version:", gatenlp.__version__)
from gatenlp import Document
GateNLP version: 1.0.8.dev3
Create a document from some text/string and print it:
doc1 = Document("This is a small test document")
print(doc1)
Document(This is a small test document,features=Features({}),anns=[])
Documents¶
In a notebook, documents are visualized using the html-viewer when a document is the last value of a cell or when display(doc1) or when document.show() is used:
# from IPython.display import display
doc1
Documents¶
The show() method can be used to influence and parametrize the viewer
doc1.show(doc_style="color: blue; font-weight: bold;")
Documents: load¶
- to load documents use
Document.load(some_location, ...) - document format is auto-detected from the extension or specified using the
fmtparameter - one standard format for saving/loading GateNLP is "bdocjs" (a JSON serialization)
some_locationcan be file or URL
doc2 = Document.load('./data/document-testing.txt')
doc2
Documents: save (JSON)¶
- use
thedocument.save(location, ...) - format is inferred from the extension or specified using
fmtparameter - Formats:
bdocjs(JSON, default),bdocym(YAML, slow),bdocmp(MessagePack, compact)
doc1.save("myfirstdocument.bdocjs")
with open("myfirstdocument.bdocjs", "rt", encoding="utf-8") as infp:
print(infp.read())
{"annotation_sets": {}, "text": "This is a small test document", "features": {}, "offset_type": "p", "name": ""}
Document: save (YAML)¶
doc1.save("myfirstdocument.bdocym") # use YAML serialization
with open("myfirstdocument.bdocym", "rt", encoding="utf-8") as infp:
print(infp.read())
annotation_sets: {}
features: {}
name: ''
offset_type: p
text: This is a small test document
# Can also "save" to memory/string, here the format is needed!
doc1.save_mem(fmt="bdocjs")
'{"annotation_sets": {}, "text": "This is a small test document", "features": {}, "offset_type": "p", "name": ""}'
Document features¶
- Documents can have arbitrary features (similar to Python dictionaries)
- key/name (string) maps to some value
- value should be JSON serializable
- name starting with single underscore: "private value"
- name starting with double underscore: "private/transient value" (not saved by default, not shown in viewer)
import datetime
doc1.features["loading_date"] = str(datetime.datetime.now())
doc1.features["purpose"] = "Testing gatenlp."
doc1.features["numeric_value"] = 22
doc1.features["dict_of_objects"] = {"dict_key": "dict_value", "a_list": [1,2,3,4,5]}
doc1.features["_tmp1"] = "some value"
doc1.features["__tmp2"] = 12345
doc1
print("1:", doc1.features["purpose"])
print("2:", doc1.features.get("doesntexist"))
print("3:", doc1.features.get("doesntexist", "NA!"))
1: Testing gatenlp. 2: None 3: NA!
Features: API¶
for name, value in doc1.features.items():
print(f"{name}: {value}")
loading_date: 2022-07-02 21:48:56.801623
purpose: Testing gatenlp.
numeric_value: 22
dict_of_objects: {'dict_key': 'dict_value', 'a_list': [1, 2, 3, 4, 5]}
_tmp1: some value
__tmp2: 12345
Features: serialization¶
Lets check how the document with features is serialized to "bdocjs" (JSON) format:
import pprint, json
js_str = doc1.save_mem(fmt="bdocjs")
js = json.loads(js_str)
pprint.pprint(js)
{'annotation_sets': {},
'features': {'_tmp1': 'some value',
'dict_of_objects': {'a_list': [1, 2, 3, 4, 5],
'dict_key': 'dict_value'},
'loading_date': '2022-07-02 21:48:56.801623',
'numeric_value': 22,
'purpose': 'Testing gatenlp.'},
'name': '',
'offset_type': 'p',
'text': 'This is a small test document'}
Annotations & Annotation Sets & Spans¶
- Span: a range of offsets
- Annotation: information about a range of offsets, has
- annotation type
- features
- unique integer annotation id
- Annotation set: named collection of annotations
- "set": only one annotation per set with the same annotation id
- but ordered by insertion order or offset
- "default" annotation set has name "" (empty string)
Adding annotations¶
- first get the annotation set we want to add the annotation to
- then create the annotation using the
addmethod of the set
# create and get an annotation set with the name "Set1"
annset = doc1.annset("Set1")
#Now, add an annotation, this method returns the newly created annotation
annset.add(0,4,"AnnType1")
Annotation(0,4,AnnType1,features=Features({}),id=0)
Annotations¶
- The annotation covers the characters 0, 1, 2, and 3, a text of length 4 (to - from = len)
- the "to" offset is the offset after the last covered character
- in Python ALL unicode code points are represented by 1 character
- In Java: UTF-16 code units
- -> Offsets different between Java and Python!
Annotations¶
# add a few more
annset.add(0, 4, "Token", {"id": "token1'"})
annset.add(5, 7, "Token", {"id": "token2'"})
annset.add(8, 9, "Token", {"id": "token3'"})
annset.add(10, 15, "Token", {"id": "token4'"})
annset.add(16, 20, "Token", {"id": "token5"})
annset.add(21, 29, "Token", {"id": "token6"})
annset.add(0, 29, "Sentence", {"what": "The first 'sentence' annotation"});
for ann in annset:
print(ann)
Annotation(0,4,AnnType1,features=Features({}),id=0)
Annotation(0,4,Token,features=Features({'id': "token1'"}),id=1)
Annotation(0,29,Sentence,features=Features({'what': "The first 'sentence' annotation"}),id=7)
Annotation(5,7,Token,features=Features({'id': "token2'"}),id=2)
Annotation(8,9,Token,features=Features({'id': "token3'"}),id=3)
Annotation(10,15,Token,features=Features({'id': "token4'"}),id=4)
Annotation(16,20,Token,features=Features({'id': 'token5'}),id=5)
Annotation(21,29,Token,features=Features({'id': 'token6'}),id=6)
Annotations: document viewer¶
doc1.show(preselect=[("Set1", ["AnnType1", "Sentence"])])
- show all annotations for a type by clicking the type name
- clicking annotation shows annotation features instead of document features
- clicking "Document" shows the document features again
- when multiple annotations overlap, need to select first which to view
Annotations/sets: remove¶
ann0 = annset.get(0) # get by annotation id
print("Annotation id=0:", ann0)
annset.remove(ann0) # remove the annotation with the annotation id of ann1
ann1 = annset.get(1)
print("Annotation id=1:", ann1)
annset.remove(1) # remove the annotation with the given id
annset.remove([2,3,4]) # remove a whole list of annotations
print("After some anns removed ", annset)
annset.clear()
print("After set cleared: ", annset)
doc1.remove_annset("Set1")
Annotation id=0: Annotation(0,4,AnnType1,features=Features({}),id=0)
Annotation id=1: Annotation(0,4,Token,features=Features({'id': "token1'"}),id=1)
After some anns removed AnnotationSet([Annotation(0,29,Sentence,features=Features({'what': "The first 'sentence' annotation"}),id=7), Annotation(16,20,Token,features=Features({'id': 'token5'}),id=5), Annotation(21,29,Token,features=Features({'id': 'token6'}),id=6)])
After set cleared: AnnotationSet([])
Annotation Relations¶
- Annotations can overlap arbitrarily
- Annotation API has methods to check how they relate to each other
- overlap, within, covering, before, after, rightoverlapping, startingat, endingwith, coextensive ...
- Annotation API implements ordering by start offset and annotation id
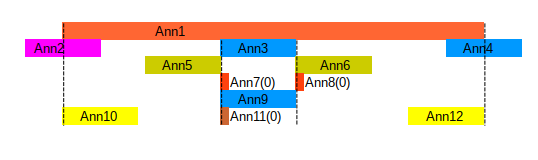
- Ann1 overlaps with all others, covers all but Ann2 and Ann4
- Ann5 is directly before Ann3, is before Ann6
- Ann10 starts at Ann1, Ann12 ends with Ann1, Ann3 and Ann9 are coextensive
doc3 = Document.load("data/ann-relations.bdocjs")
doc3.show(htmlid="view1")
Annotation Relations API¶

# make a variable for each annotation type
for anntype in list(doc3.annset("set1").type_names):
vars()[anntype.lower()] = doc3.annset("set1").with_type(anntype).for_idx(0)
print("Ann2 isoverlapping Ann1:", ann2.isoverlapping(ann1))
print("Ann2 isbefore Ann3:", ann2.isbefore(ann3))
print("Ann3 isafter Ann2:", ann3.isafter(ann2))
print("Ann1 iscovering Ann5:", ann1.iscovering(ann5))
print("Ann3 iscoextensive Ann9:", ann3.iscoextensive(ann9))
print("Ann6 iswithin Ann1:", ann6.iswithin(ann1))
print("Ann4 isrightoverlapping Ann1:", ann4.isrightoverlapping(ann1))
Ann2 isoverlapping Ann1: True Ann2 isbefore Ann3: True Ann3 isafter Ann2: True Ann1 iscovering Ann5: True Ann3 iscoextensive Ann9: True Ann6 iswithin Ann1: True Ann4 isrightoverlapping Ann1: True
Spans¶
- Objects that describe offset ranges
- similar API for relations
- can get from annotations, use when only the span of an annotation is needed
from gatenlp import Span
span1 = Span(3,4)
span2 = ann2.span
span3 = doc3.annset("set1").span
span4 = Span(ann5)
print([f"span{i}: {s}" for i, s in enumerate([span1, span2, span3, span4])])
['span0: Span(3,4)', 'span1: Span(0,6)', 'span2: Span(0,45)', 'span3: Span(12,18)']
AnnotationSet: retrieve by relation¶
- get all annotations that overlap/are before/start at/... an annotation/span/annotation set
- returns a new annotation set
- returned set is detached: not part of document, changes do set not affect document
- returned set is initially immutable: set cannot be changed
- but annotations are mutable and still the same as in the set!
- possible to "detach" annotations by (deep)copying them
set1 = doc3.annset("set1") # "attached" set
print("Within Ann1: ", [a.type for a in set1.within(ann1)])
print("Coextensive with Ann3:", [a.type for a in set1.coextensive(ann3)])
print("Coextensive with span of Ann3:", [a.type for a in set1.coextensive(ann3.span)])
Within Ann1: ['Ann10', 'Ann5', 'Ann3', 'Ann7', 'Ann9', 'Ann11', 'Ann6', 'Ann8', 'Ann12'] Coextensive with Ann3: ['Ann9'] Coextensive with span of Ann3: ['Ann3', 'Ann9']
AnnotationSet: detached / immutable¶
print("Size of set1:", len(set1))
subset1 = set1.within(ann1)
print("Size of subset1:", len(subset1))
Size of set1: 12 Size of subset1: 9
# try to add an annotation to subset1:
try:
subset1.add(2,3,"ANewOne")
except Exception as ex:
print("Got exception:", ex)
Got exception: Cannot add an annotation to an immutable annotation set
AnnotationSet: detached / immutable¶
# make the set mutable and try again
subset1.immutable = False
subset1.add(2,3,"ANewOne")
print("Size of set1:", len(set1))
print("Size of subset1:", len(subset1))
print("Is set1 detached:", set1.isdetached())
print("Is subset1 detached:", subset1.isdetached())
Size of set1: 12 Size of subset1: 10 Is set1 detached: False Is subset1 detached: True
- annotation only got added to
subset1, NOT the original set - detached sets cannot get attached again
- annotations in the detached set are the same as in the document, so changing their features will affect the document!
- detached set can also detach its annotations using
subset1.clone_anns()
Document loading/saving¶
Supported formats:
- bdocjs, bdocym, bdocmp: load/save (aliasing: only bdocym)
- GATE xml: load (but only basic data types, no aliasing)
- HTML: load and create annotations for HTML entities
- plain text: load / save
- tweet: load v1 format, WIP!
- pickle: load/save
- html-ann-viewer: save (also used for displaying in jupyter)
Document: load HTML¶
# lets load and view the main GateNLP documentation page:
doc4 = Document.load("https://gatenlp.github.io/python-gatenlp/", fmt="html")
doc4
Document: view sets/types¶
Use: doc.show(annspec=["set1", ("set2", "type1"), ("set3", ["type1", "type2"])]
doc4.show(annspec=[("Original markups", ["h1","h2","a","li"])])
Document: save html-ann-viewer¶
doc4.save("gatenlp-doc.html", fmt="html-ann-viewer", notebook=False, stretch_height=True)
from IPython.display import IFrame
IFrame("gatenlp-doc.html", 900,400)
Exchange Documents with Java GATE¶
- Python GateNLP can read Java GATE XML format
- GATE plugin Format_Bdoc provides support for loading/saving formats bdocjs, bdocym and bdocmp in Java GATE
- Offsets differ between GATE and GateNLP:
- Java: offsets refer to UTF-16 encoding, possibly a surrogate pair of UTF-16 characters
- Python: offsets refer to Unicode code points
- bdocjs/bdocym/bdocmp automatically convert the offsets on either side
- field
offset_typeis eitherporj
Corpus¶
- a list-like collection of a fixed number of documents which can be retrieved and stored by index:
get:doc = corpus[2]set:corpus[3] = doc - on retrieval, the index gets stored in a document feature
- implements
store(doc)to save a document to the index stored in the document feature - some implementations:
append(doc)to add a new document to the corpus - some implementations: store/retrieve
None- on retrieveal:
Noneindicates absence of document - on storing:
Noneindicates that document should get removed or should not get updated
- on retrieveal:
ListCorpus¶
- wrap a Python list-like data structure
- but provide the
storemethod
from gatenlp.corpora import ListCorpus
texts = ["this is text one", "here is text two", "and this is text three"]
docs = [Document(t) for t in texts]
lcorp = ListCorpus(docs)
doc1 = lcorp[1]
print(doc1.features)
lcorp.store(doc1)
Features({'__idx_139956274540624': 1})
DirFilesCorpus¶
- all (recursive) files in a directory with some specific extension
- specify some specific format or infer from file extension
- stores the relative file path as a document feature
from gatenlp.corpora import DirFilesCorpus
corp1 = DirFilesCorpus("data/dir1") # get all the matching filenames from the directory
print("Number of documents:", len(corp1))
doc1 = corp1[2] # actually read the document from the directory
print("Text for idx=2:", doc1.text)
print("Features for idx=2:", doc1.features)
doc1.annset().add(0,len(doc1.text), "Document", dict(what="test document"))
# this writes the document back to the file:
corp1.store(doc1)
# could also have used: corp1[2] = doc1
Number of documents: 4
Text for idx=2: This is another document for testing which mentions John Smith.
Features for idx=2: Features({'__idx_139955830010000': 2})
Corpus Viewer¶
from gatenlp.visualization import CorpusViewer
cviewer = CorpusViewer(corp1)
cviewer.show()
Other Corpus Classes¶
NumberedDirFilesCorpus: create a directory tree where the path represents digits of a large number- e.g.
000/002/341.bdocfor element number 2341 of 600000000 total
- e.g.
EveryNthCorpus: wrap a corpus and access only elements $k*i + o$ for $i = 0..\lfloor(n/k)\rfloor$- $k$: every that many elements
- $o$: start with this element ($o < k$)
- e.g.: get elements 3, 7, 11, 15 from a corpus with 17 elements
- useful for processing files in a DirFilesCorpus with multiple processes
ShuffledCorpus: random re-ordering of the elements in the wrapped corpusCachedCorpus: store retrieved elements from a (slow) base corpus in a (fast) cache corpus- Still work in progress
Source, Destination¶
- Document Source: something that can be iterated over to get one Document after the other
- unknown size
- a Corpus may also function as a Source
- Document Destination: something that has
append(doc)to add Document instances- unknown final size
- also has
close()to end writing - may implement the
with documentdestination as dest:pattern - an appendable Corpus may also function as a Destination
Source, Destination examples¶
BdocjsLinesFileSource/Destination: one line of bdocjs serialization per documentTsvFileSource: one column in a TSV file contains the text, other columns can be stored in featuresPandasDfSource: similar to TSV source, but for a Pandas data frame- Still work in progress: improvements/more to come!
TsvFileSource¶
from gatenlp.corpora import TsvFileSource
tsvsrc1 = TsvFileSource("data/mytsvfile.tsv", text_col="text", feature_cols=dict(src="source",year="year"))
for doc in tsvsrc1:
print(doc)
Document(This is the text of the first row. It has several sentences.,features=Features({'src': 'source1', 'year': '2005'}),anns=[])
Document(Text of the second row.,features=Features({'src': 'source1', 'year': '2006'}),anns=[])
Document(Another text, this time of the third row. ,features=Features({'src': 'source2', 'year': '2001'}),anns=[])
Document(And here another, from the fourth row.,features=Features({'src': 'source3', 'year': '2013'}),anns=[])
PandasDfSource¶
from gatenlp.corpora import PandasDfSource
try: # this requires Pandas!
import pandas as pd, csv
df = pd.read_csv("data/mytsvfile.tsv", sep="\t", quotechar=None, index_col=None, quoting=csv.QUOTE_NONE)
pdsrc1 = PandasDfSource(df, text_col="text", data_cols=["source", "year"])
for doc in pdsrc1:
print(doc)
except:
print("Pandas not installed")
Document(This is the text of the first row. It has several sentences.,features=Features({'__data': {'source': 'source1', 'year': 2005}}),anns=[])
Document(Text of the second row.,features=Features({'__data': {'source': 'source1', 'year': 2006}}),anns=[])
Document(Another text, this time of the third row. ,features=Features({'__data': {'source': 'source2', 'year': 2001}}),anns=[])
Document(And here another, from the fourth row.,features=Features({'__data': {'source': 'source3', 'year': 2013}}),anns=[])
Conll-U Source¶
- Read in one of the many multilingual corpora from https://universaldependencies.org/
- create documents from k sentences, paragraphs conll documents
- use original text hints or space hints, if available
- Example: first few lines of
ar-ud-train.conllu
from gatenlp.corpora.conll import ConllUFileSource
src = ConllUFileSource("data/ar-tiny.conllu", group_by="doc", group_by_n=1)
corp = list(src)
print(len(corp))
3
Conll-U Source¶
corp[0].show(doc_style="direction: rtl; font-size: 1.5em; line-height: 1.5;")
Annotators, Executors¶
- Annotator: a callable that accepts a document to process and either:
- returns the same or a different document (most common situation)
- returns None: something went wrong or the document should get filtered
- returns a list of zero to n documents: filter, error, split documents
- may be just a function, but usually a subclass of
Annotator - standard methods for handling over-a-corpus results
- Pipeline: a special annotator that recursively runs other annotators in sequence
- Executor: a class that runs an annotator
- on a corpus
- on a source and optional destination
- takes care of handling None, lists of returned documents
Example 1/3¶
from gatenlp.corpora import ListCorpus
from gatenlp.processing.pipeline import Pipeline
from gatenlp.processing.annotator import AnnotatorFunction
from gatenlp.processing.executor import SerialCorpusExecutor
texts = ["Some text.", "Another text.", "Also some text here.", "And this is also some text."]
docs = [Document(t) for t in texts]
corp = ListCorpus(docs)
def annfunc1(doc):
doc.annset().add(0,3,"Ann1")
return doc
def annfunc2(doc):
doc.annset("set1").add(1,4,"Type1")
return doc
ann1 = AnnotatorFunction(annfunc1)
ann2 = AnnotatorFunction(annfunc2)
pipeline = Pipeline()
pipeline.add(ann1, name="FirstAnnotator")
pipeline.add(ann2, name="SecondAnnotator")
Example 2/3¶
exe = SerialCorpusExecutor(pipeline, corpus=corp)
exe()
corp[2]
Example 3/3¶
# use corp as source and create another ListCorpus as destination
corpnew = ListCorpus([])
exe2 = SerialCorpusExecutor(pipeline, source=corp, destination=corpnew)
exe2()
print("Length of corpnew:", len(corpnew))
print(f"in={exe2.n_in}, out={exe2.n_out}, none={exe2.n_none}, ok={exe2.n_ok}, err={exe2.n_err}")
corpnew[2]
Length of corpnew: 4 in=4, out=4, none=0, ok=4, err=0
Spacy Annotator¶
- Use a SpaCy pipeline to annotate a document
- convert spacy tokens, entities etc into Annotations, convert token attributes into annotation features
- makes it much easier to add own annotations and features, no need to keep vocab files around
- but possibly not as optimized/fast as Spacy
Preparation:
- make sure spacy dependency is installed for your gatenlp environment:
pip install -U spacy(not necessary ifgatenlp[all]was used) - make sure the model for the language is installed:
English:python -m spacy download en_core_web_sm - To use in notebook, need to restart kernel after installation!
Spacy Annotator¶
import spacy
print("Spacy version:", spacy.__version__)
from gatenlp.lib_spacy import AnnSpacy
nlp = spacy.load("en_core_web_sm")
annotator = AnnSpacy(pipeline=nlp, outsetname="Spacy")
doc2.annset("Spacy").clear() # avoid annotation duplication when running several times
doc2 = annotator(doc2)
/home/johann/software/anaconda/envs/gatenlp-37/lib/python3.7/site-packages/torch/cuda/__init__.py:80: UserWarning: CUDA initialization: The NVIDIA driver on your system is too old (found version 9010). Please update your GPU driver by downloading and installing a new version from the URL: http://www.nvidia.com/Download/index.aspx Alternatively, go to: https://pytorch.org to install a PyTorch version that has been compiled with your version of the CUDA driver. (Triggered internally at ../c10/cuda/CUDAFunctions.cpp:112.) return torch._C._cuda_getDeviceCount() > 0
Spacy version: 3.3.1
/home/johann/software/anaconda/envs/gatenlp-37/lib/python3.7/site-packages/spacy/util.py:837: UserWarning: [W095] Model 'en_core_web_sm' (3.2.0) was trained with spaCy v3.2 and may not be 100% compatible with the current version (3.3.1). If you see errors or degraded performance, download a newer compatible model or retrain your custom model with the current spaCy version. For more details and available updates, run: python -m spacy validate warnings.warn(warn_msg)
Spacy Annotator¶
# Adapt size of viewer
from IPython.core.display import display, HTML
display(HTML("<style>#view2-wrapper { font-size: 80% !important; } #view2-row1 {height: 15em; min-height: 5em;}</style>"))
doc2.show(htmlid="view2")
Stanza Annotator¶
- Use a Stanza pipeline to annotate a document
- convert stanza tokens, entities etc into Annotations, convert token attributes into annotation features
Preparation:
- make sure stanza dependency is installed for your gatenlp environment:
pip install -U stanza(not necessary ifgatenlp[all]was used) - make sure the model for the language is installed:
English:python -c "import stanza; stanza.download('en')"
Stanza Annotator¶
import stanza
print("Stanza version:",stanza.__version__)
from gatenlp.lib_stanza import AnnStanza
nlpstanza = stanza.Pipeline(logging_level="ERROR")
annotatorstanza = AnnStanza(pipeline=nlpstanza, outsetname="Stanza")
doc2.annset("Stanza").clear() # avoid annotation duplication when running several times
doc2 = annotatorstanza(doc2)
Stanza version: 1.3.0
Stanza Annotator¶
# change size of document viewer
from IPython.core.display import display, HTML
display(HTML("<style>#view3-wrapper { font-size: 100% !important; } #view3-row1 {height: 10em; min-height: 5em;}</style>"))
doc2.show(htmlid="view3")
Gazetteers¶
- Look up tokens/words/strings/phrases in a long list ("gazetteer list")
- StringGazetteer: match document text against list of known strings
- TokenGazetter:
- Match sequences of tokens
- Tokens in documents: based on sequences of annotations
- Use underlying document text or some feature value per annotation ("Token")
- gazetteer list: each entry should be a list of tokens as well!
StringGazetteer: gazetteer list¶
from gatenlp.processing.gazetteer import StringGazetteer
# Strings we want to find
sgazlist1 = [
("Barack Obama", dict(url="https://en.wikipedia.org/wiki/Barack_Obama")),
("Obama", dict(url="https://en.wikipedia.org/wiki/Barack_Obama")),
("Donald Trump", dict(url="https://en.wikipedia.org/wiki/Donald_Trump")),
("Trump", dict(url="https://en.wikipedia.org/wiki/Donald_Trump")),
("George W. Bush", dict(url="https://en.wikipedia.org/wiki/George_W._Bush")),
("George Bush", dict(url="https://en.wikipedia.org/wiki/George_W._Bush")),
("Bush", dict(url="https://en.wikipedia.org/wiki/George_W._Bush")),
(" Bill Clinton ", dict(url="https://en.wikipedia.org/wiki/Bill_Clinton")),
("Clinton", dict(url="https://en.wikipedia.org/wiki/Bill_Clinton")),
]
StringGazetteer: document¶
# Document with some text mentioning some of the names in the gazeteer for testing
text = """Barack Obama was the 44th president of the US and he followed George W. Bush and
was followed by Donald Trump. Before Bush, Bill Clinton was president.
Also, lets include a sentence about South Korea which is called 대한민국 in Korean.
And a sentence with the full name of Iran in Farsi: جمهوری اسلامی ایران and also with
just the word "Iran" in Farsi: ایران
Also barack obama in all lower case and SOUTH KOREA in all upper case
"""
doc0 = Document(text)
doc0
StringGazetteer: match¶
sgaz1 = StringGazetteer(source=sgazlist1, source_fmt="gazlist")
doc0 = sgaz1(doc0)
doc0
Document Tokenization¶
- Stanza and Spacy both produce Tokens
- Also possible to use NLTKTokenizer: wrap any of the NLTK tokenizers
from gatenlp.processing.tokenizer import NLTKTokenizer
from nltk.tokenize.destructive import NLTKWordTokenizer # get some tokenizer to use
nltk_tokenizer = NLTKTokenizer(nltk_tokenizer=NLTKWordTokenizer(), token_type="Token")
doc2.annset().clear()
doc2 = nltk_tokenizer(doc2)
Document Tokenization¶
doc2
TokenGazetteer: list¶
1) Use prepared list with already tokenized entries
from gatenlp.processing.gazetteer import TokenGazetteer
gazlist1 = [
(["Donald", "Trump"], dict(what="person", country="US")),
(["Boris", "Johnson"], dict(what="person", country="UK")),
(["Google"], dict(what="company", country="Everywhere, really!"))
]
tgaz1 = TokenGazetteer(source=gazlist1, source_fmt="gazlist",
annset_name="", outset_name="TGaz1", ann_type="Lookup")
doc2 = tgaz1(doc2)
TokenGazetteer: match¶
doc2 = tgaz1(doc2)
doc2
TokenGazetteer: listfile¶
2) Load a list from a file, using JAVA GATE "def" format (https://gate.ac.uk/userguide/sec:annie:gazetteer):
- A somthing.def file contains one line for each list file to use
- Each somename.lst file contains one line with entry to match and arbitrary features
E.g. data/gaz1.def:
persons.lst
companies.lstdata/persons.lst:
Donald Trump what=person country=US
Boris Johnson what=person country=UKdata/companies.lst:
Google where=Everywhere, really!Gazetteer List (GATE def)¶
tgaz2 = TokenGazetteer(
source="data/gaz1.def", source_fmt="gate-def", annset_name="", outset_name="TGaz2",
ann_type="Lookup", source_tokenizer=nltk_tokenizer)
doc2.annset("TGaz2").clear()
doc2 = tgaz2(doc2)
Gazetteer List¶
Result when using the loaded GATE-style gazetteer files:
doc2
TokenGazetteer¶
Other features:
- Match each gazetteer token against the feature of a document token, not the underlying document text (e.g. the lemma)
- only match within another annotation type (e.g. only in noun phrases)
- More to come: still WIP!
- handle space tokens
- handle split tokens (do not cross them!)
Regexp Annotator¶
- match multiple regular expressions
- use macros to build complex REs from simpler parts
- different strategies for how to handle multiple matches
- assign annotation features from RE groups
from gatenlp.processing.gazetteer import StringRegexAnnotator
RegexpAnnotator: rules¶
rules = """
year=(19[0-9]{2}|20[0-9]{2})
month=(0[0-9]|10|11|12)
day=([012][0-9]|3[01])
// The ISO date:
|{{year}}-{{month}}-{{day}}
0 => Date type="iso", year=G1, month=G2, day=G3
# The traditional way of writing a date:
|{{day}}/({{month}})/{{year}}
0 => Date type="traditional", year=G3, month=G2, day=G1
"""
re_anntr = StringRegexAnnotator(source=rules, source_fmt="string")
RegexpAnnotator: match¶
redoc = Document("""
A document that contains a date here: 2013-01-12 and also here: 14/02/1991. This should not
get matched: 1833-12-21 and nor should this 45/03/2012 but this should 13/12/2012 and also
this, despite not being a valid data: 31/02/2000
""")
re_anntr(redoc)
redoc
PAMPAC¶
- PAttern Matching through PArser Combinators
- A pattern language for matching annotations and text
- DSL (domain specific language) implemented as Python classes
- Purpose: similar to Java GATE JAPE / JAPE Plus, but:
- does not need a separate language
- more powerful (e.g. directly match text AND annotations)
PAMPAC - How to use¶
- define Rules
- each Rule consists of:
- a Pattern to match in the document
- an Action to perform if the pattern matches
- create a Pampac instance from the rules
- run the Pampac instance on the document, using a set of annotations and specify the output annotation set
PAMPAC - Example 1¶
Lets create a rule that annotates any Token which is within a PERSON or ORG annotation:
from gatenlp.pam.pampac import Ann, AnnAt, Rule, Pampac, AddAnn, N, Seq, Or
from gatenlp.pam.matcher import FeatureMatcher, IfNot
r1 = Rule(
# first the pattern
Or ( Ann("Token", name="tok").within("ORG"),
Ann("Token", name="tok").within("PERSON")
),
# then the action for the pattern
AddAnn(name="tok", type="PersOrOrg")
)
# get the annotations we want to use for matching
anns2match = doc2.annset("Stanza").with_type(["Token", "PERSON", "ORG"])
outset = doc2.annset("Pampac1")
outset.clear()
# Create the Pampac instance from the single rule and run it on the annotations, also specify output set
# The run method returns the list of offsets and the action return values where the rule matches in the doc
Pampac(r1).run(doc2, anns2match, outset=outset)
len(outset)
15
PAMPAC - Example 1¶
doc2
PAMPAC - Example 2¶
Create a rule that annotates any Sequence of two or more Token annotations which have a "upos" tag of "PROPN", separated by at most one other arbitrary token:
from gatenlp.pam.pampac import Ann, AnnAt, Rule, Pampac, AddAnn, N, Seq
from gatenlp.pam.matcher import FeatureMatcher, IfNot
feat = FeatureMatcher(upos="PROPN")
r1 = Rule(
# first the pattern
Seq( Ann("Token", features=feat),
N( Seq( N(Ann("Token", features=IfNot(feat)), min=0, max=1),
Ann("Token", features=feat)),
min=1, max=99),
name="seq1"
),
# then the action for the pattern
AddAnn(name="seq1", type="PROPNSEQ")
)
# get the annotations we want to use for matching
anns2match = doc2.annset("Stanza").with_type("Token")
outset = doc2.annset("Pampac2")
outset.clear()
# Create the Pampac instance from the single rule and run it on the annotations, also specify output set
# The run method returns the list of offsets and the action return values where the rule matches in the doc
Pampac(r1).run(doc2, anns2match, outset=outset)
len(outset)
7
PAMPAC - Example 2¶
Result: found 8 matches and added annotations for them:
doc2
Processing¶
- Instance of Annotator: process a document and return it (usually the same document)
- optionally: return None or a list of documents
- Pipeline:
- run several Annotators in Sequence
- handle None / list of returned documents
- also handle per-corpus initialization, finishing, return values
Executor (work in progress!)
- Run a pipeline on a corpus or a document source / destination pair
Runner:
- run a pipeline on a corpus, source/destination pair from the command line
- Multiprocessing capable
- run several workers locally or on a cluster (using Ray infrastructure)
- corpus / source / destination must be multiprocessing capable, duplicated over nodes
- e.g. DirFilesCorpus, DirFilesSource init:
nparts=1, partnr=0
Machine Learning¶
- Currently Work in Progress
- Not part of the
gatenlppackage - Planned: for each ML/DL backend a separate package
- upcoming:
gatenlp-ml-huggingface - basic functionality working:
gatenlp-ml-tnerfor chunking / NER
- upcoming:
- Install ML package into gatenlp environment or install gatenlp into ML package environment
- Most modern ML systems require GPU / powerful computers
What about Ontologies?¶
- Java GATE has Ontology-related plugins
- but very outdated and limited (no support for OWL2)
- (Currently) no direct support in GateNLP
- But: Python packages rdflib, Owlready2 and maybe others
- Finding terms:
- Extract names, process names as docs, build gazetteer, use gazetteer to match and annotate with URI
- match classes, subclasses, etc: directly use e.g. Owlready2 API from e.g. PAMPAC or in own annotators
GATE Worker¶
- Allows running the Java GATE process from Python
- API for exchanging document and performing frequent Java GATE tasks from Python
- Py4J API to run ANY Java from Python
- Python connects to a Java process, communicates over sockets
- Option 1: Start GATE GUI, load PythonWorkerLr, then connect a GateNLP GateWorker to it
- Option 2: Start Java GATE worker using the
gatenlp-gate-workercommand - Option 3: directly start the Java GATE worker when creating the GateNLP GateWorker instance
Let's try Option 3 first: GATE_HOME environment variable must be set, or must know GATE installation directory
GATE Worker¶
from gatenlp.gateworker import GateWorker
gs = GateWorker()
# if GATE_HOME not set use gs = GateWorker(gatehome="/where/Gate/is/Installed")
# if java is not on the PATH use gs = GateWorker(java=""/path/to/the/java/binary")
# If port(s) in use e.g.: `ss -ltp -at dport=:25333`
Trying to start GATE Worker on port=25333 host=127.0.0.1 log=false keep=false Process id is 1303916
CREOLE plugin loaded: creole CREOLE plugin loaded: Format: Bdoc 1.10 Plugin Python version: 3.0.7 commit: 9adf5ed dirty: false Lib interaction version: 4.1 commit: 7819f1c dirty: false Python gatenlp version: 1.0.7 commit: 8c15d82 dirty: false CREOLE plugin loaded: Python 3.0.7
PythonWorkerRunner.java: starting server with 25333/127.0.0.1/LdhVaxhZGVsS_rbwE78Og6AFs7o/false
# Create a GATE document on the JAVA GATE side and return a handle
gdoc1 = gs.createDocument("An example document mentioning Barack Obama and New York")
# Can call Java API methods on that handle and get/convert the result
print(gdoc1.getClass())
print(gdoc1.getName())
print(gdoc1.getAnnotationSetNames())
class gate.corpora.DocumentImpl GATE Document_00015 set()
GATE Worker¶
# lets load the prepared ANNIE pipeline on the Java side and process the GATE document with it
gs.loadMavenPlugin("uk.ac.gate.plugins", "annie", "9.0")
gpipe = gs.loadPipelineFromPlugin("uk.ac.gate.plugins", "annie", "/resources/ANNIE_with_defaults.gapp")
gcorp = gs.newCorpus()
gcorp.add(gdoc1)
gpipe.setCorpus(gcorp)
gpipe.execute()
CREOLE plugin loaded: ANNIE 9.0
GATE Worker¶
So far, everything happened on the Java side, use a GateWorker API method to convert the document into a Python GateNLP document:
pdoc1 = gs.gdoc2pdoc(gdoc1)
pdoc1
GateWorker¶
- Stopping: the GateWorker (Java process) can get stopped using
gs.close() - Will also automatically stop when the Python process ends
gs.close()
Java GatenlpWorker ENDING: 1303916
GateWorker Annotator¶
An annotator to process Python GateNLP documents with a Java GATE pipeline
from gatenlp.gateworker import GateWorkerAnnotator
# Specify a prepared GATE pipeline file to get loaded into Java GATE
# Specify a GateWorker
gw = GateWorker(port=31313)
gs_app = GateWorkerAnnotator(pipeline="data/annie.xgapp", gateworker=gw)
Trying to start GATE Worker on port=31313 host=127.0.0.1 log=false keep=false Process id is 1303940
CREOLE plugin loaded: creole CREOLE plugin loaded: Format: Bdoc 1.10 Plugin Python version: 3.0.7 commit: 9adf5ed dirty: false Lib interaction version: 4.1 commit: 7819f1c dirty: false Python gatenlp version: 1.0.7 commit: 8c15d82 dirty: false CREOLE plugin loaded: Python 3.0.7
PythonWorkerRunner.java: starting server with 31313/127.0.0.1/hLZUII_z566eMAyEUH5pVCZdDig/false
CREOLE plugin loaded: ANNIE 9.0
GateWorkerAnnotator¶
Example, running on a directory corpus:
from gatenlp.processing.executor import SerialCorpusExecutor
dircorpus = DirFilesCorpus("data/dir1", sort=True)
exe = SerialCorpusExecutor(annotator=gs_app, corpus=dircorpus)
exe()
gw.close()
tmpdoc = dircorpus[2]
print(tmpdoc.features)
tmpdoc
Features({'gate.SourceURL': 'created from String', '__idx_139952273354960': 2})
Java GatenlpWorker ENDING: 1303940
Java GATE Python Plugin¶
- Java GATE Plugin
- Documentation: http://gatenlp.github.io/gateplugin-Python/
- provides a Processing Resource (PR) PythonPr
- PythonPr: makes it possible to use a Python program to annotated GATE documents
- Plugin comes with its own copy of the
gatenlppackage!- but can use
gatenlpinstalled separately as well
- but can use
Requirements¶
- Python 3.6 or higher installed
- possibly own Python environment
conda create -n gatenlp python=3.9
sortedcontainerspackagepip install sortedcontainers
- Optionally other dependencies for using Spacy etc.
Using the Plugin in GATE¶
- Requires GATE 8.6.1 or later
- Load from Plugin Manager
- make sure to use latest version
- make sure "Load always" was not checked, NOT already loaded!
- make sure "Save session" is not enabled, if necessary restart!
- Or: will get loaded with a pipeline that uses the plugin automatically
Create new Pipeline¶
- Load the Python plugin: check "Load Now" for "Python", then click "Apply All"
- Create PR PythonPr: right click "Processing Resource", choose "PythonPr"
- Click file dialog button for the
pythonPrograminit parameter - In the file selection dialog, choose directory and enter a non-existing name
test1.py, click Open, then OK - When the specified file does not exist, it is created with a template content
- Double click PR to show in GATE
- Save often!
Edit PythonPr Program¶
- from the GATE GUI:
- Save: write content to the file (no warning if changed with other program!)
- Save & Use: also check for syntax error
- with external editor:
- changes NOT automatically reflected in GATE Editor!
- to use a changed file: right click PythonPr and select "Reinitialise"
PythonPr Program Template¶
from gatenlp import Document, AnnotationSet, GateNlpPr, interact
@GateNlpPr
class MyAnnotator:
# the following method is run on every document, this method must exist:
def __call__(self, doc, **kwargs):
pass
# the start and finish methods are optional, if they exist the start
# method is called before the first document of a corpus and the finish
# method is called after the last document.
# def start(self, **kwargs):
# pass
# def finish(self, **kwargs):
# pass
# THE FOLLOWING MUST BE PRESENT SO THAT GATE CAN COMMUNICATE WITH THE PYTHON PROCESS!
if __name__ == "___main__": # NOTE: changed from __main__ to ___main__ to prevent running in Notebook!
interact()
PythonPr Program Example¶
from gatenlp import Document, AnnotationSet, GateNlpPr, interact
@GateNlpPr
class MyAnnotator:
def __init__(self):
self.n_docs = 0
def __call__(self, doc, **kwargs):
self.n_docs += 1
doc.annset().add(0,3,"SomeType")
doc.features["docnr"] = self.n_docs
def start(self, **kwargs):
print("Processing starting, we got kwargs:", kwargs)
self.n_docs = 0
def finish(self, **kwargs):
print("Processing finished, documents processed: ", self.n_docs)
if __name__ == "___main__": # NOTE: changed from __main__ to ___main__ to prevent running in Notebook!
interact()
Create PythonPr Pipeline¶
- new Application (Pipeline)
- add PythonPr to the pipeline
- Review Runtime Parameters
- Create/load a document
- Create a corpus for the document
- Double-click pipeline, choose corpus
- Run Application
- See example pipeline
pythonpr-example1.xgapp
PythonPr Runtime Parameters¶
pythonBinary/pythonBinaryUrl: ifpythonis not on the path or you want to use a specific binary- use a specific environment by using the binary from that environment!
usePluginGatenlpPackage: the Python plugin contains its own version of Pythongatenlp, iffalseuse whatever is installed into the environment insteadsetsToUse: replace*with a list of Annotation Set names to avoid transferring lots of existing anntoationsprogramParams: send arbitrary parameters to the Python program (as kwargs)- can get pre-set if a JSON file
pythonscript.py.parmsexists
- can get pre-set if a JSON file
configFile: select any file/dir to pass as_config_fileparameter to the Python program (as kwarg)
How it works¶
- when pipeline/contoller runs, a separate Python process is started
- the Python code is loaded
- communication is done via pipes over stdin/stdout between Java and Python
interact()communicates with the Java PythonPr- when the pipeline starts, the
startmethod is called,programParamspassed on - for each document:
- the document is converted to bdoc json, transferred, converted to GateNLP document and passed to
__call__ - the changes to the document done via the gatenlp API are recorded and sent back to Java
- PythonPr applies the changes to the document
- the document is converted to bdoc json, transferred, converted to GateNLP document and passed to
- when the pipeline finishes, the
finishmethod is called and any results returned to Java
Multiprocessing¶
- In GATE, multiprocessing is done via duplication of the pipeline and running pipelines in parallel
- Done via gcp, easily using
gcp-direct.sh - PythonPr supports duplication and multiprocessing
- One parallel Python process for each duplicate
- number of duplicates and duplicate id passed to each process
- if more than one duplicate, the
reduce(resultslist)method is invoked - see example pipeline
pythonpr-count-words.xgapp
PythonPr Prepared Pipelines¶
- Two prepared pipelines: for running Spacy and Stanza
- Right click Applications - Ready Made Applications - Python
- python-spacy
- python-stanford-stanza
What to use when?¶
- Java GATE "Classic":
- need existing plugins/pipelines, need GUI editing/annotation
- within Java app; GCP / Mimir involved
- Gate Worker from Python:
- Need Java GATE plugins/pipelines
- but Python context / application
- consider running in Java GATE first, process result files with Python GateNLP
- Python GateNLP:
- Python context, ML/DNN (Pytorch, Tensorflow), numpy, ...
- Want to use Stanza/Spacy; GateNLP-only functions
More documentation:
- Python GateNLP https://gatenlp.github.io/python-gatenlp/
- Java GATE Python Plugin: http://gatenlp.github.io/gateplugin-Python/
- Java GATE Format_Bdoc plugin: https://gatenlp.github.io/gateplugin-Format_Bdoc/
Thank You & Have Fun with GATE and GateNLP!¶